Generates plots comparing predictions with observations.
pred.plot(pred, obs, ...)
# S3 method for default
pred.plot(
pred,
obs,
xlab = "Predicted",
ylab = "Observed",
lm.fit = TRUE,
lowess = TRUE,
...
)
# S3 method for factor
pred.plot(
pred,
obs,
type = c("frec", "perc", "cperc"),
xlab = "Observed",
ylab = NULL,
legend.title = "Predicted",
label.bars = TRUE,
...
)Arguments
- pred
a numeric vector with the predicted values.
- obs
a numeric vector with the observed values.
- ...
additional graphical parameters or further arguments passed to other methods (e.g. to
RcmdrMisc::Barplot()).- xlab
a title for the x axis.
- ylab
a title for the y axis.
- lm.fit
logical indicating if a
lmfit is added to the plot.- lowess
logical indicating if a
lowesssmooth is added to the plot.- type
types of the desired plots. Any combination of the following values is possible:
"frec"for frequencies,"perc"for percentages or"cperc"for conditional percentages.- legend.title
a title for the legend.
- label.bars
if
TRUE(the default) show values of frequencies or percents in the bars.
Value
The default method invisibly returns the fitted linear model if
lm.fit == TRUE.
pred.plot.factor() invisibly returns the horizontal coordinates of the
centers of the bars.
Details
The default method draws a scatter plot of the observed values against the predicted values.
pred.plot.factor() creates bar plots representing frequencies, percentages
or conditional percentages of pred within levels of obs.
This method is a front end to RcmdrMisc::Barplot().
See also
Examples
set.seed(1)
nobs <- nrow(hbat)
itrain <- sample(nobs, 0.8 * nobs)
train <- hbat[itrain, ]
test <- hbat[-itrain, ]
# Regression
fit <- lm(fidelida ~ velocida + calidadp, data = train)
pred <- predict(fit, newdata = test)
obs <- test$fidelida
res <- pred.plot(pred, obs)
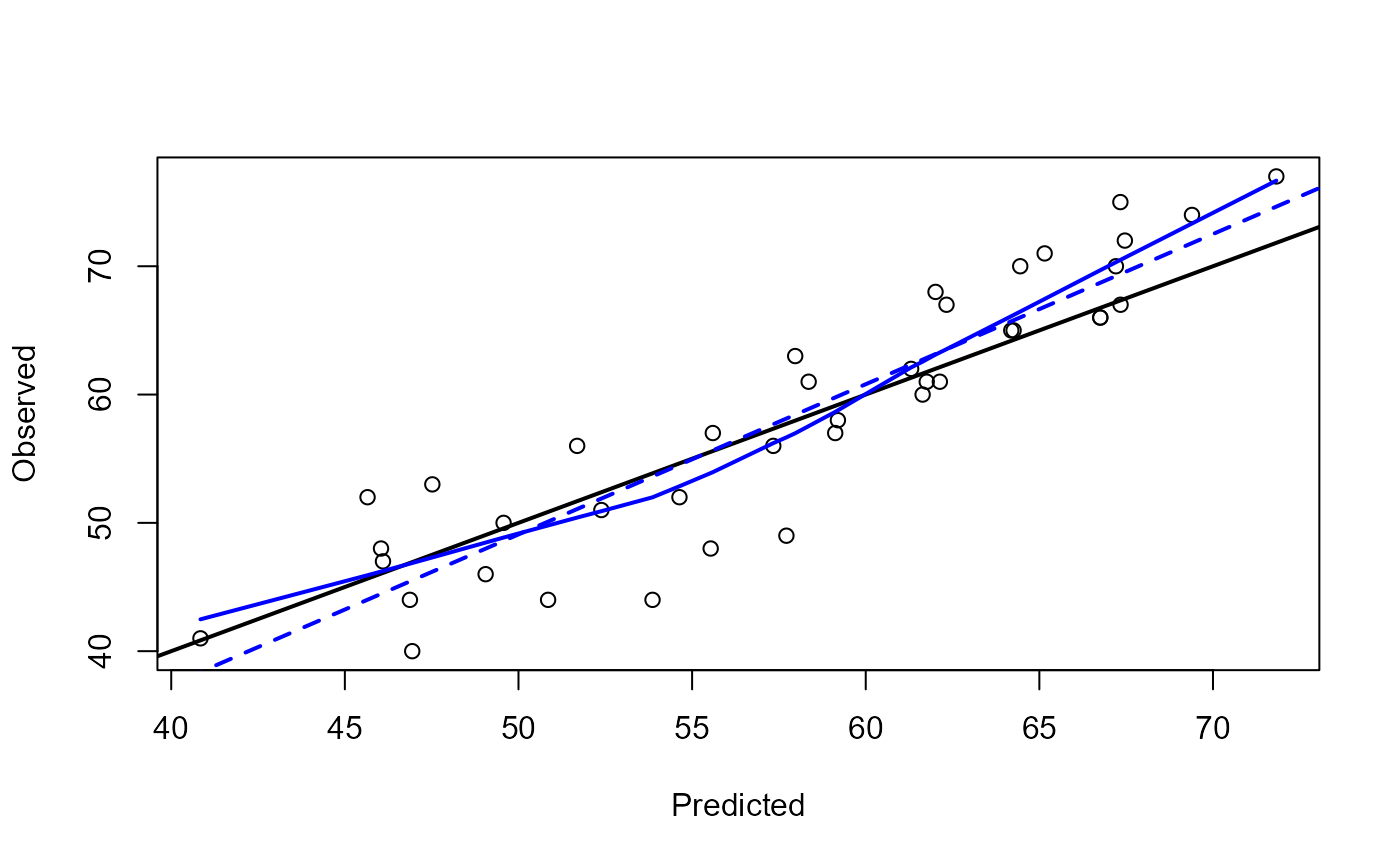 summary(res)
#>
#> Call:
#> lm(formula = obs ~ pred)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -9.6170 -2.4273 -0.5227 2.8353 7.9903
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -9.43398 4.92829 -1.914 0.0631 .
#> pred 1.17064 0.08433 13.881 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 4.206 on 38 degrees of freedom
#> Multiple R-squared: 0.8353, Adjusted R-squared: 0.8309
#> F-statistic: 192.7 on 1 and 38 DF, p-value: < 2.2e-16
#>
# Classification
fit2 <- glm(alianza ~ velocida + calidadp, family = binomial, data = train)
obs <- test$alianza
p.est <- predict(fit2, type = "response", newdata = test)
pred <- factor(p.est > 0.5, labels = levels(obs))
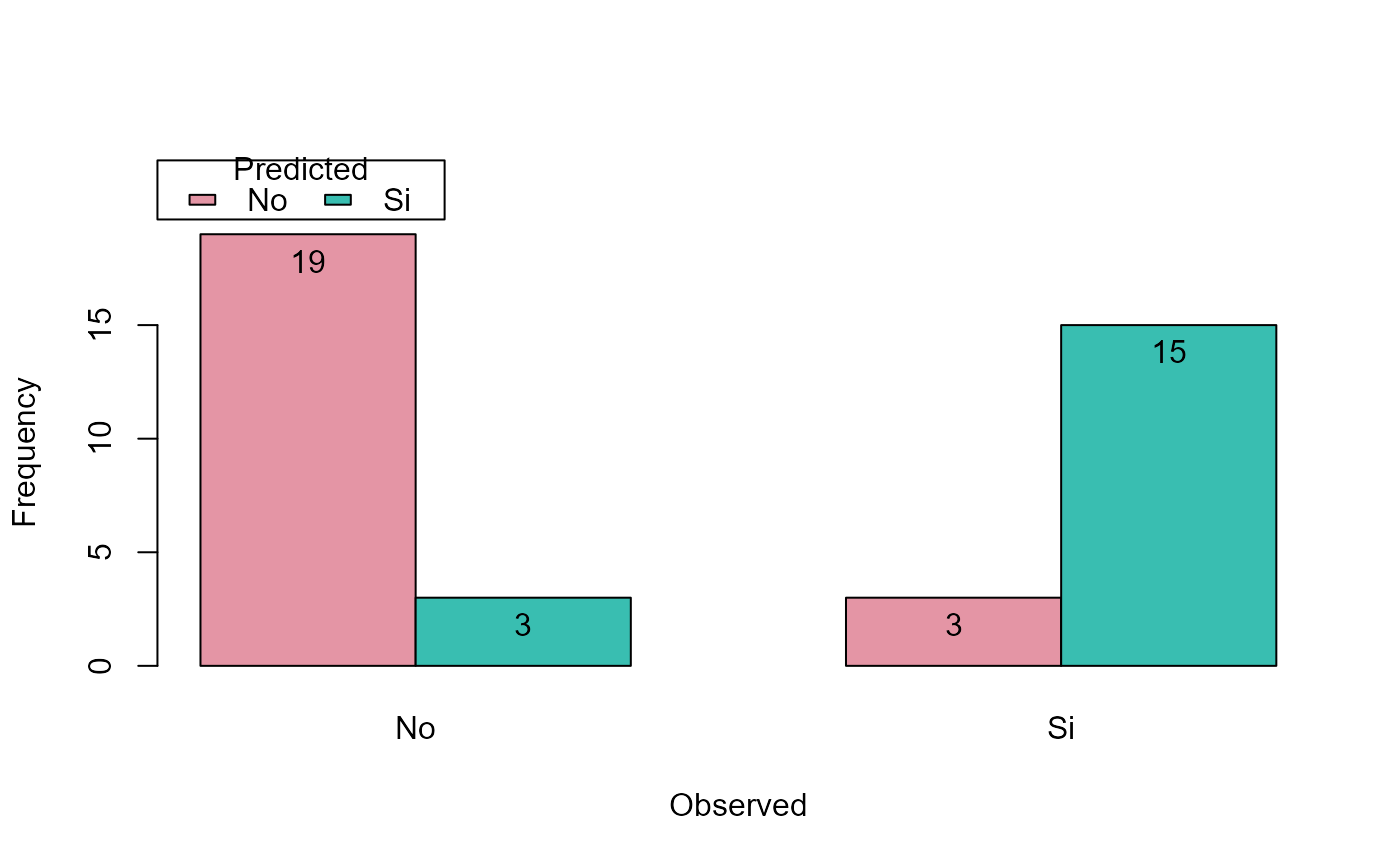
pred.plot(pred, obs, type = "frec", style = "parallel")
summary(res)
#>
#> Call:
#> lm(formula = obs ~ pred)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -9.6170 -2.4273 -0.5227 2.8353 7.9903
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -9.43398 4.92829 -1.914 0.0631 .
#> pred 1.17064 0.08433 13.881 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 4.206 on 38 degrees of freedom
#> Multiple R-squared: 0.8353, Adjusted R-squared: 0.8309
#> F-statistic: 192.7 on 1 and 38 DF, p-value: < 2.2e-16
#>
# Classification
fit2 <- glm(alianza ~ velocida + calidadp, family = binomial, data = train)
obs <- test$alianza
p.est <- predict(fit2, type = "response", newdata = test)
pred <- factor(p.est > 0.5, labels = levels(obs))
pred.plot(pred, obs, type = "frec", style = "parallel")
 old.par <- par(mfrow = c(1, 2))
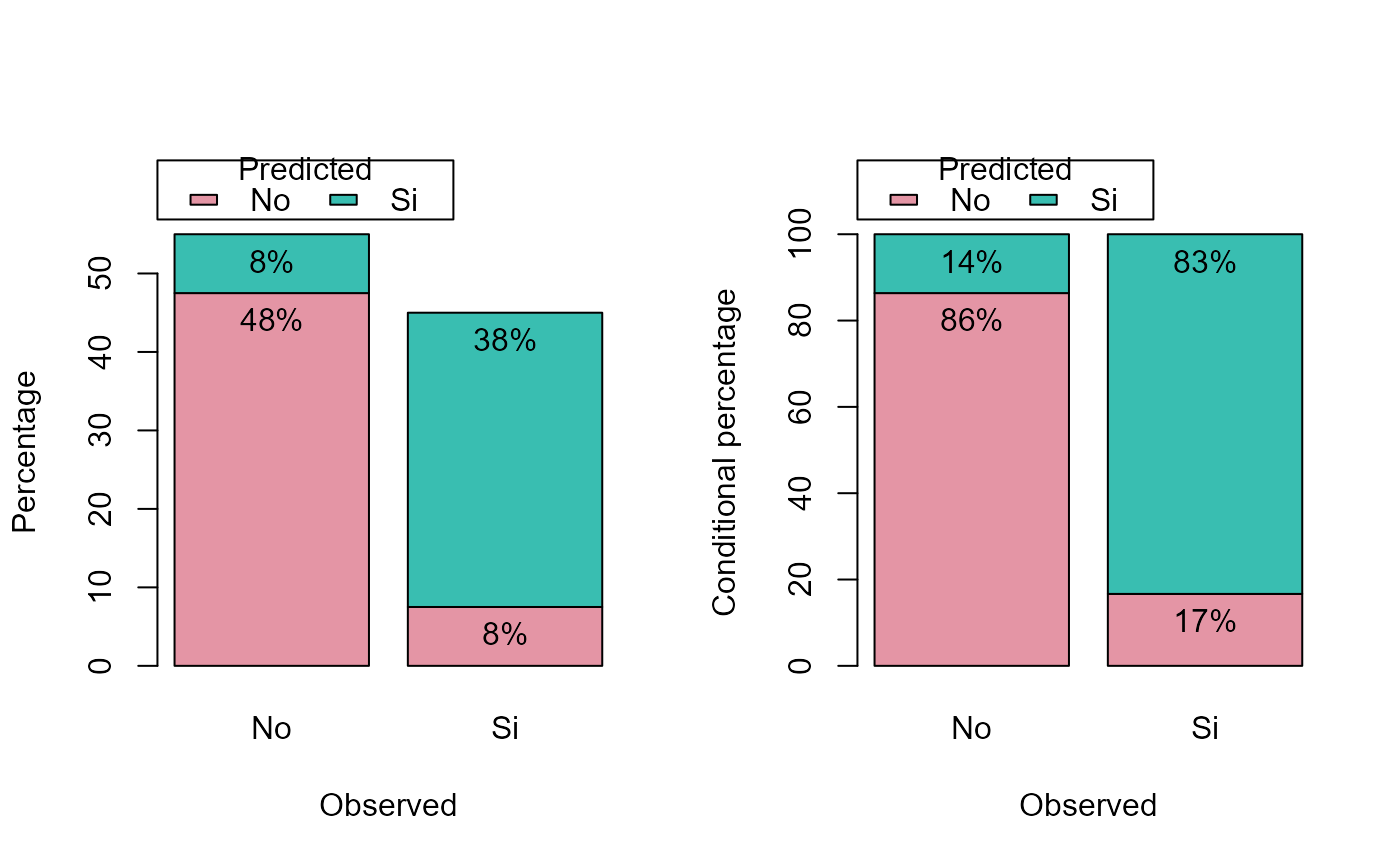
pred.plot(pred, obs, type = c("perc", "cperc"))
old.par <- par(mfrow = c(1, 2))
pred.plot(pred, obs, type = c("perc", "cperc"))
 par(old.par)
par(old.par)