Estimates the functional mean (and its first derivatives) using local polynomial kernel smoothing.
# S3 method for npf.data
locpol(
x,
h = NULL,
degree = 1 + as.numeric(drv),
drv = FALSE,
hat.bin = FALSE,
ncv = 0,
...
)
# S3 method for npf.bin
locpol(
x,
h = NULL,
degree = 1 + as.numeric(drv),
drv = FALSE,
hat.bin = FALSE,
ncv = 0,
...
)
# S3 method for npf.locpol
predict(object, newx = NULL, ...)
# S3 method for npf.locpol
residuals(object, var, as.npf.data = TRUE, ...)
# S3 method for npf.locpol
plot(
x,
y = NULL,
main = deparse(substitute(x)),
col = "lightgray",
legend = FALSE,
...
)Arguments
- x
an object (of class
npf.dataornpf.bin) used to select a method.- h
(full) bandwidth matrix (controls the degree of smoothing; only the upper triangular part of h is used).
- degree
degree of the local polynomial used. Defaults to 1 (local linear estimation).
- drv
logical; if
TRUE, the matrix of estimated first derivatives is returned.- hat.bin
logical; if
TRUE, the hat matrix of the binned data is returned.- ncv
integer; determines the number of cells leaved out in each dimension. Defaults to 0 (the full data is used) and it is not normally changed by the user in this setting. See "Details" below.
- ...
additional graphical parameters (passed to
plot.npf.data).- object
object used to select a method.
- newx
vector with the (irregular) points to predict (interpolate).
- var
(optional) a vector or an object of class
npf.varwith the estimated (or theoretical) variances.- as.npf.data
logical; if
TRUE(default) returns anpf.data-class object containing the residuals.- y
(optional) a vector or an object of class
npf.varwith the estimated (or theoretical) variances.- main
plot title.
- col
color table used to set up the color scale (see
imagefor details).- legend
logical; if
TRUE(default), the plotting region is splitted into two parts, drawing the main plot in one and the legend with the color scale in the other. IfFALSEonly the (coloured) main plot is drawn and the arguments related to the legend are ignored (npsp::splot()is not called).
Value
locpol.npf.data and locpol.npf.bin return an S3 object of class
npf.locpol extending npsp::locpol.bin (locpol + bin data + grid par.).
If newx == NULL, predict.npf.locpol returns the trend estimates
corresponding to the discretization points
(otherwise npsp::interp.data.grid is called).
residuals.npf.locpol returns the residuals, standardized if var
is not missing, and as a npf.data-class object if as.npf.data = TRUE.
See also
Examples
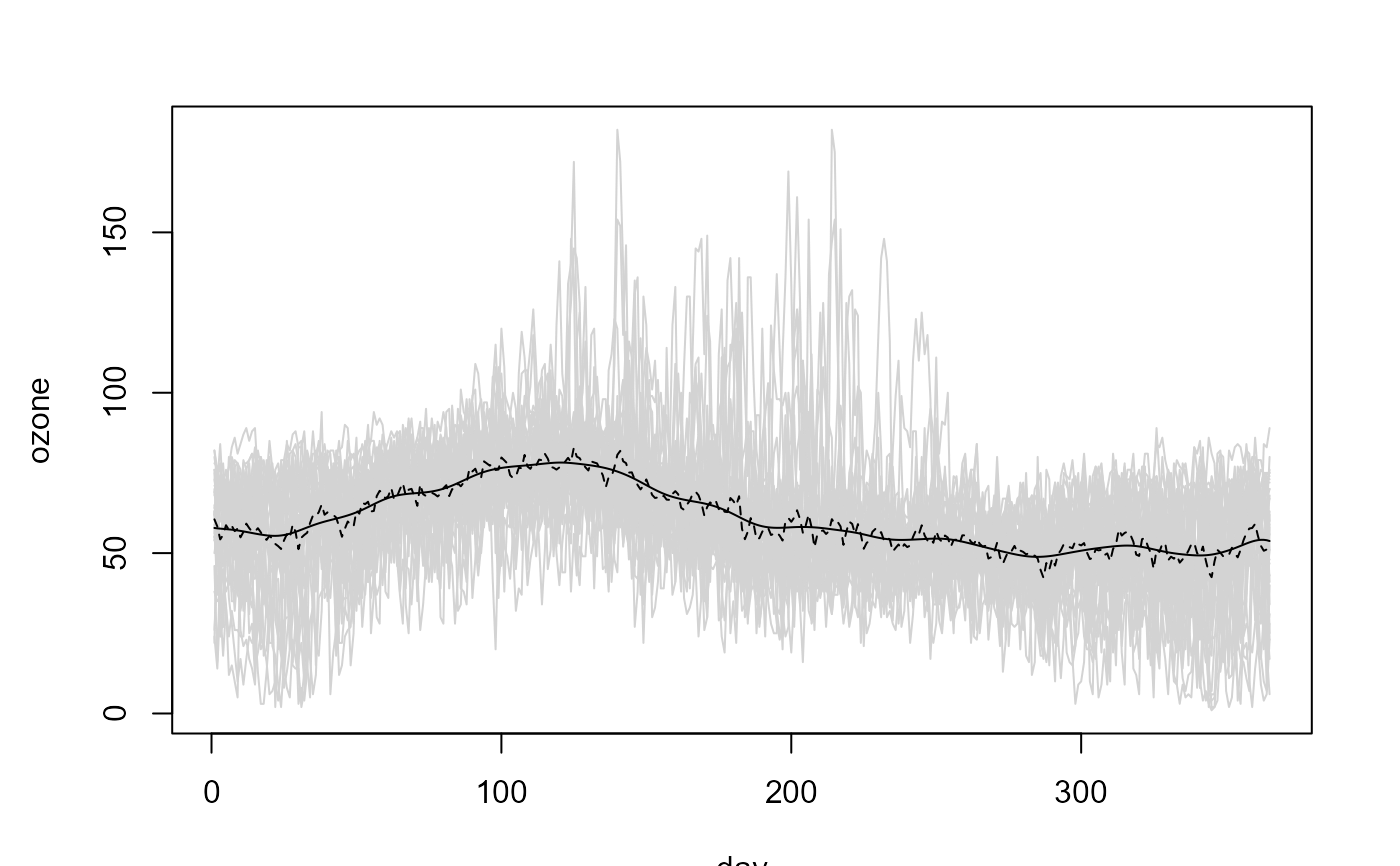
fd <- npf.data(ozone, dimnames = "day")
# Linear Local trend estimate
lp <- locpol(fd, h = 35)
# Plot
plot(fd, col = "lightgray", legend = FALSE)
lines(lp$data$x, lp$biny, lty = 2) # x = coords(fd)
lines(lp$data$x, lp$est)
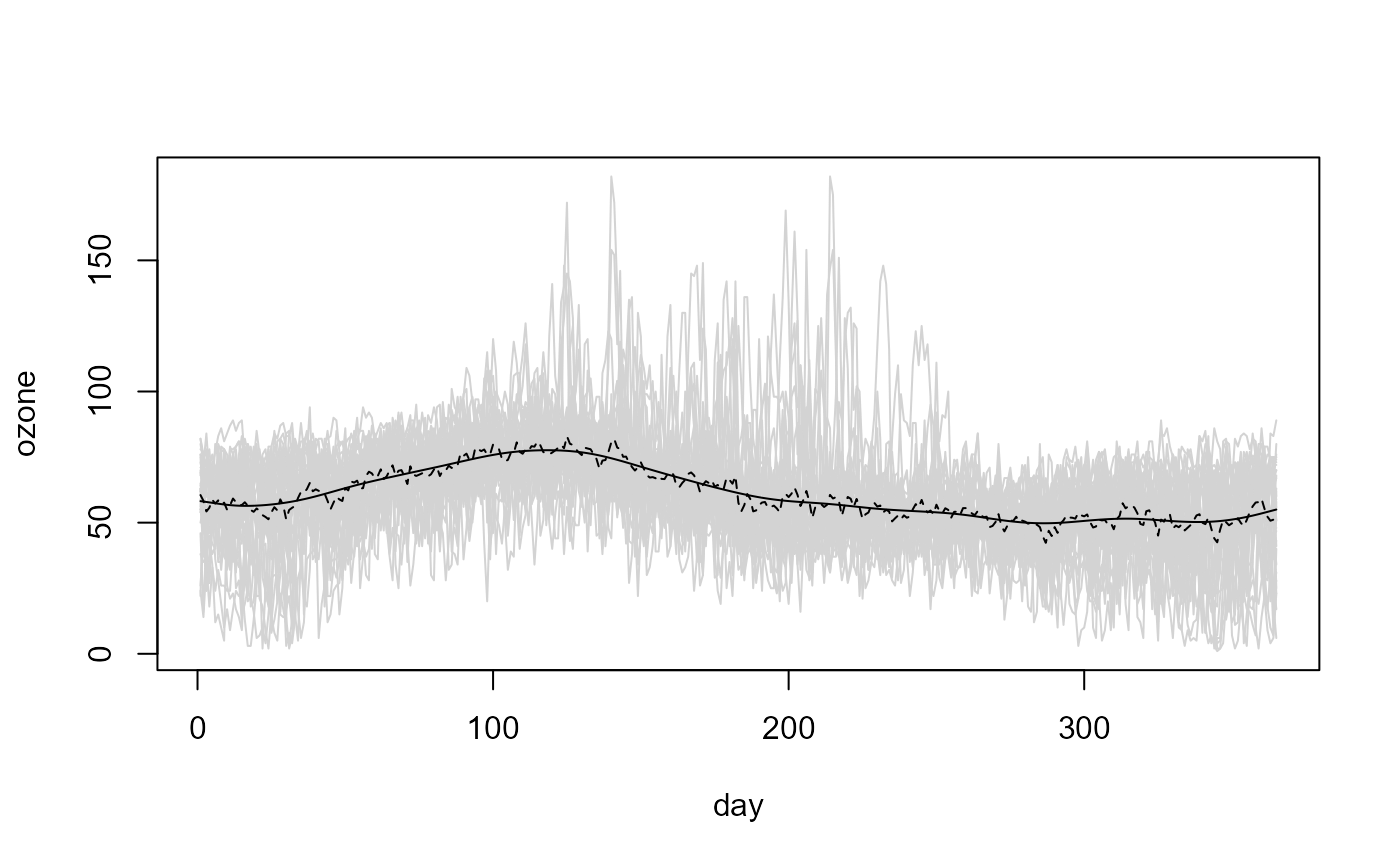 # Bandwidth selection
# (assuming independence and homoscedasticity)
bin <- npf.binning(fd) # binning
trend.h <- h.cv(bin)$h
trend.h
#> [,1]
#> [1,] 4.947342
# the selected bandwidth undersmoothes the data...
trend.h <- 4*trend.h
# Linear Local trend estimate
lp <- locpol(bin, h = trend.h, hat.bin = TRUE)
# Plot
plot(fd, col = "lightgray", legend = FALSE)
lines(lp$data$x, lp$biny, lty = 2)
lines(lp$data$x, lp$est)
# Bandwidth selection
# (assuming independence and homoscedasticity)
bin <- npf.binning(fd) # binning
trend.h <- h.cv(bin)$h
trend.h
#> [,1]
#> [1,] 4.947342
# the selected bandwidth undersmoothes the data...
trend.h <- 4*trend.h
# Linear Local trend estimate
lp <- locpol(bin, h = trend.h, hat.bin = TRUE)
# Plot
plot(fd, col = "lightgray", legend = FALSE)
lines(lp$data$x, lp$biny, lty = 2)
lines(lp$data$x, lp$est)