splot is designed to combine a standard R plot with
a legend representing a (continuous) color scale. This is done by splitting
the plotting region into two parts. Keeping one for the main chart and
putting the legend in the other.
For instance, sxxxx functions (spoints, simage
and spersp) draw the corresponding high-level plot (xxxx),
after calling splot, to include a legend strip for the color scale.
These functions are based on function image.plot of package
fields, see its documentation for additional information.
jet.colors and hot.colors create a color table useful for contiguous
color scales and scolor assigns colors to a numerical vector.
splot(
slim = c(0, 1),
col = jet.colors(128),
breaks = NULL,
horizontal = FALSE,
legend.shrink = 0.9,
legend.width = 1.2,
legend.mar = ifelse(horizontal, 3.1, 5.1),
legend.lab = NULL,
bigplot = NULL,
smallplot = NULL,
lab.breaks = NULL,
axis.args = NULL,
legend.args = NULL,
add = FALSE
)
scolor(s, col = jet.colors(128), slim = range(s, finite = TRUE))
jet.colors(n)
hot.colors(n, rev = TRUE)Arguments
- slim
limits used to set up the color scale.
- col
color table used to set up the color scale (see
imagefor details).- breaks
(optional) numeric vector with the breakpoints for the color scale: must have one more breakpoint than
coland be in increasing order.- horizontal
logical; if
FALSE(default) legend will be a vertical strip on the right side. IfTRUEthe legend strip will be along the bottom.- legend.shrink
amount to shrink the size of legend relative to the full height or width of the plot.
- legend.width
width in characters of the legend strip. Default is 1.2, a little bigger that the width of a character.
- legend.mar
width in characters of legend margin that has the axis. Default is 5.1 for a vertical legend and 3.1 for a horizontal legend.
- legend.lab
label for the axis of the color legend. Default is no label as this is usual evident from the plot title.
- bigplot
plot coordinates for main plot. If not passed these will be determined within the function.
- smallplot
plot coordinates for legend strip. If not passed these will be determined within the function.
- lab.breaks
if breaks are supplied these are text string labels to put at each break value. This is intended to label axis on a transformed scale such as logs.
- axis.args
additional arguments for the axis function used to create the legend axis (see
image.plotfor details).- legend.args
arguments for a complete specification of the legend label. This is in the form of list and is just passed to the
mtextfunction. Usually this will not be needed (seeimage.plotfor details).- add
logical; if
TRUEthe legend strip is just added to the existing plot (the graphical parameters are not changed).- s
values to be converted to the color scale.
- n
number of colors (
>= 1) to be in the palette.- rev
logical; if
TRUE, the palette is reversed (decreasing overall luminosity).
Value
splot invisibly returns a list with the following 3 components:
- bigplot
plot coordinates of the main plot. These values may be useful for drawing a plot without the legend that is the same size as the plots with legends.
- smallplot
plot coordinates of the secondary plot (legend strip).
- old.par
previous graphical parameters (
par(old.par)will reset plot parameters to the values before entering the function).
jet.colors and hot.colors return a character vector of colors (similar to
heat.colors or terrain.colors; see rgb).
Details
scolor converts a real valued vector to a color scale. The range
slim is divided into length(col) + 1 pieces of equal length.
Values which fall outside the range of the scale are coded as NA.
jet.colors generates a rainbow style color table similar to the MATLAB (TM)
jet color scheme. It may be appropriate to distinguish between values above and
below a central value (e.g. between positive and negative values).
hot.colors generates a color table similar to the MATLAB (TM)
hot color scheme (reversed by default). It may be appropriate to represent values
ranging from 0 to some maximum level (e.g. density estimation).
The default value rev = TRUE may be adecuate to grayscale convertion.
Side Effects
After exiting splot, the plotting region may be changed
(par("plt")) to make it possible to add more features to the plot.
See also
spoints, simage, spersp,
image, image.plot.
Examples
# Plot equivalent to spoints():
scale.range <- range(aquifer$head)
res <- splot(slim = scale.range)
with( aquifer, plot(lon, lat, col = scolor(head, slim = scale.range),
pch = 16, cex = 1.5, main = "Wolfcamp aquifer data"))
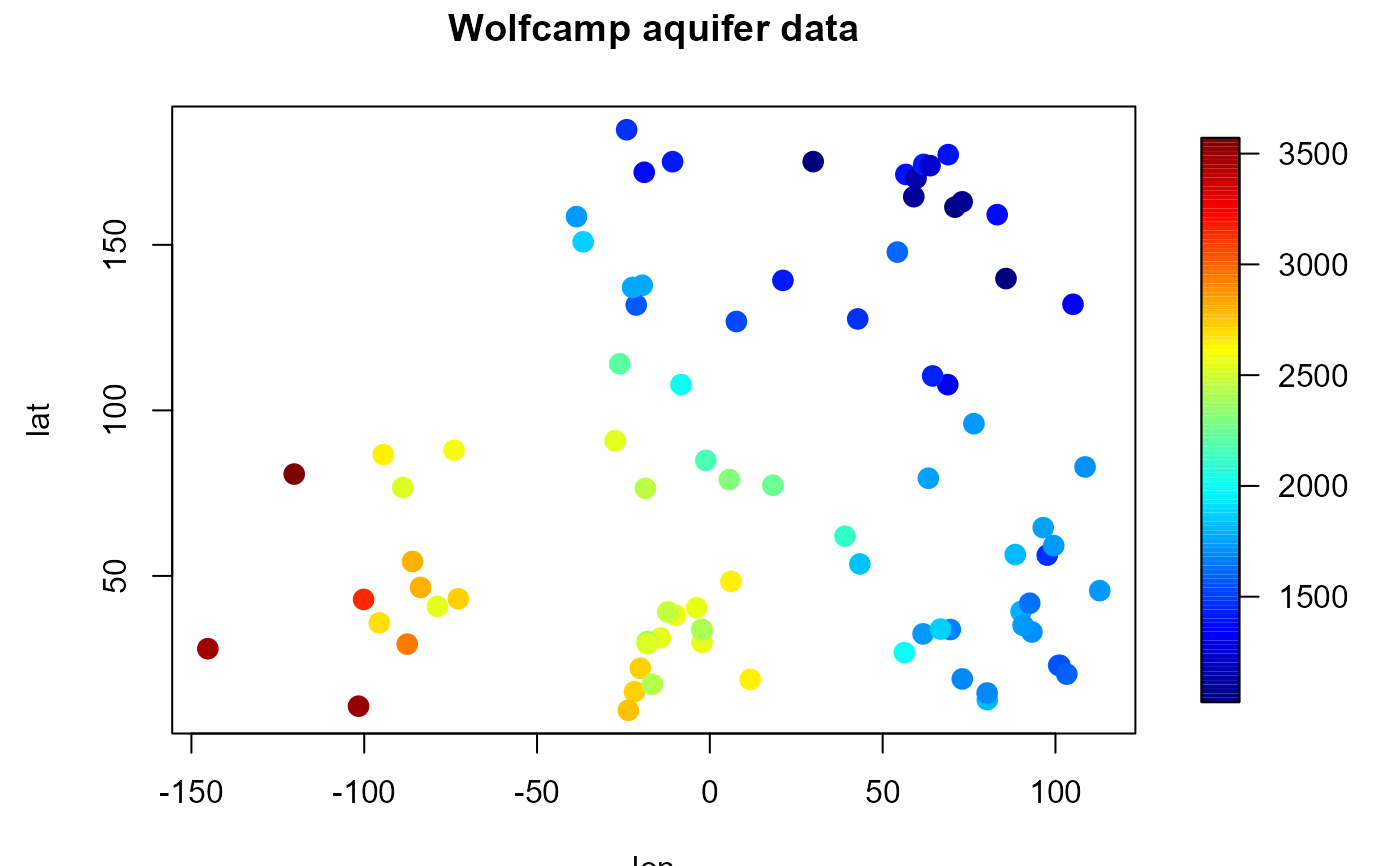 par(res$old.par) # restore graphical parameters
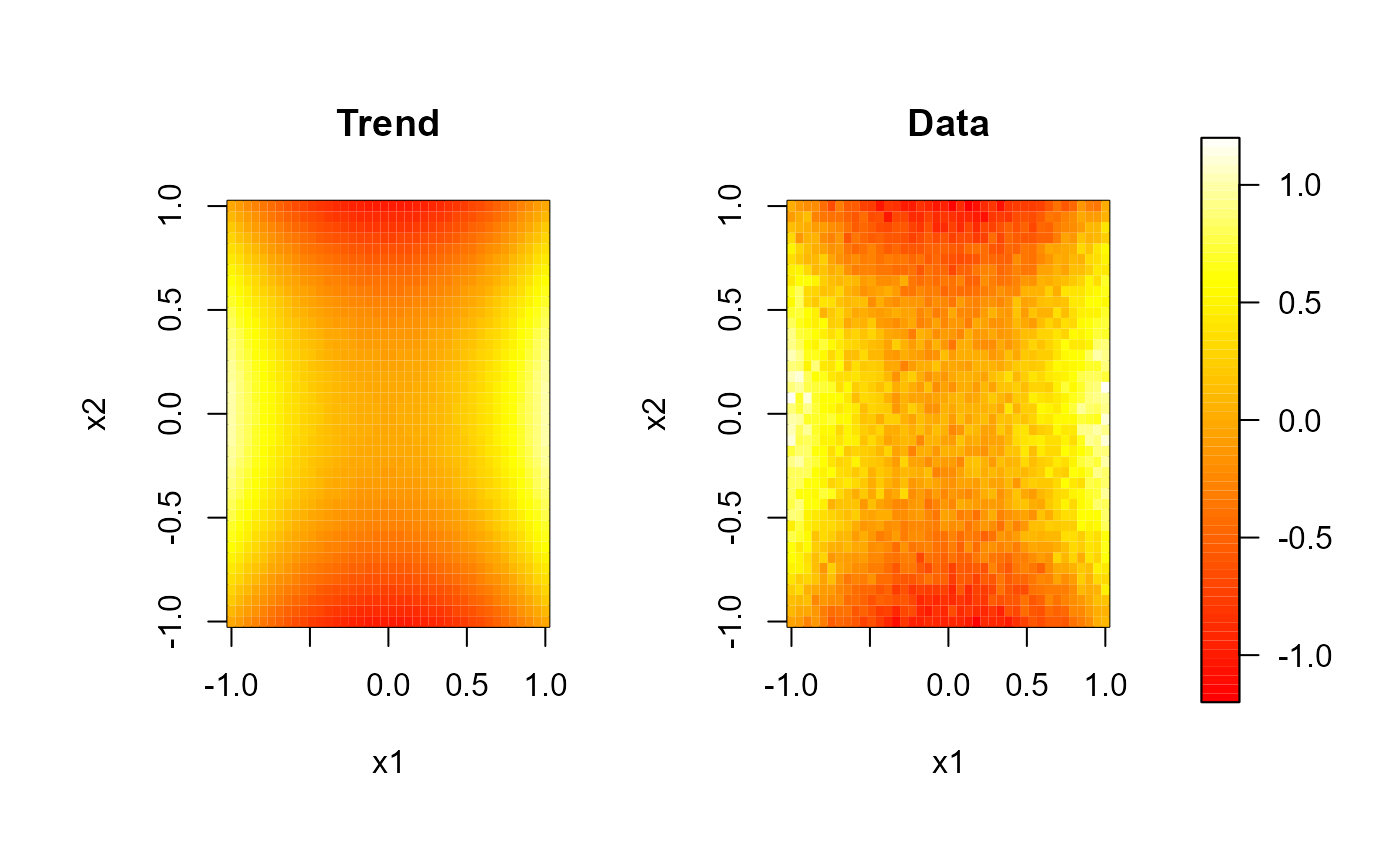
# Multiple plots with a common legend:
# regularly spaced 2D data...
set.seed(1)
nx <- c(40, 40) # ndata = prod(nx)
x1 <- seq(-1, 1, length.out = nx[1])
x2 <- seq(-1, 1, length.out = nx[2])
trend <- outer(x1, x2, function(x,y) x^2 - y^2)
y <- trend + rnorm(prod(nx), 0, 0.1)
scale.range <- c(-1.2, 1.2)
scale.color <- heat.colors(64)
# 1x2 plot with some room for the legend...
old.par <- par(mfrow = c(1,2), omd = c(0.05, 0.85, 0.05, 0.95))
image( x1, x2, trend, zlim = scale.range, main = 'Trend', col = scale.color)
image( x1, x2, y, zlim = scale.range, main = 'Data', col = scale.color)
par(old.par)
# the legend can be added to any plot...
splot(slim = scale.range, col = scale.color, add = TRUE)
par(res$old.par) # restore graphical parameters
# Multiple plots with a common legend:
# regularly spaced 2D data...
set.seed(1)
nx <- c(40, 40) # ndata = prod(nx)
x1 <- seq(-1, 1, length.out = nx[1])
x2 <- seq(-1, 1, length.out = nx[2])
trend <- outer(x1, x2, function(x,y) x^2 - y^2)
y <- trend + rnorm(prod(nx), 0, 0.1)
scale.range <- c(-1.2, 1.2)
scale.color <- heat.colors(64)
# 1x2 plot with some room for the legend...
old.par <- par(mfrow = c(1,2), omd = c(0.05, 0.85, 0.05, 0.95))
image( x1, x2, trend, zlim = scale.range, main = 'Trend', col = scale.color)
image( x1, x2, y, zlim = scale.range, main = 'Data', col = scale.color)
par(old.par)
# the legend can be added to any plot...
splot(slim = scale.range, col = scale.color, add = TRUE)
 ## note that argument 'zlim' in 'image' corresponds with 'slim' in 'sxxxx' functions.
## note that argument 'zlim' in 'image' corresponds with 'slim' in 'sxxxx' functions.